Return of Genomic Results and Estimating Penetrance in Population-Based Cohorts
Genome sequencing (GS) is increasingly conducted in large-scale human research studies, creating questions for investigators about whether and how to return genetic findings. The American College of Medical Genetics and Genomics (ACMG) recommends that physicians report secondary findings in at least 59 actionable genes (the ACMG59) whenever clinical GS is ordered. In an effort to address ethical concerns of investigators and the expectations of participants, ACMG recommendations are being applied by many investigators to guide genomic return of results (gRoR) in research biobanks and longitudinal studies. Critical knowledge gaps exist about the benefits and harms of gRoR on research participants and the impact on investigators, how to efficiently identify and confirm such variants, and how to provide accurate estimates about their penetrance, particularly among participants in population-based studies and in underrepresented minorities. Empirical data on these questions are urgently needed to guide protocol development for research participants sequenced by the NHLBI Trans-Omics for Precision Medicine (TOPMed) Program and to inform the All of Us Research Program (AoURP) within the Precision Medicine Initiative, which plans to offer 1 million diverse participants access to their genomic data and to some interpreted results.
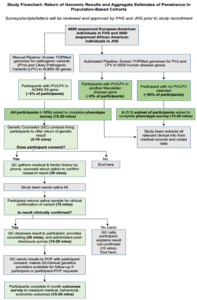
Recently 7603 individuals in the Framingham Heart Study (FHS) and the Jackson Heart Study (JHS) were sequenced as part of TOPMed, of whom 2885 and 2674 (total of 5559) respectively are alive. PopSeq aims to return genomic information in these populations and begin the gROR with the ACMG59. The study will collect outcomes data that will lay the groundwork for return of results for variants in additional genes. It will generate crucial data for other population studies and biobanks, will inform future efforts to scale the labor-intensive process of variant classification, and will allow testing of a new method to begin to estimate penetrance among populations unselected for family history.
The specific aims of this project are to (1) return clinically actionable genomic results to participants and track outcomes among the living participants, (2) improve high-throughput methods for identifying valid pathogenic variation, and (3) evaluate aggregate penetrance for Mendelian diseases.
Although there is increasing interest in the return of genomic research results from large population- based studies, there are currently no standard procedures or guidelines for doing so and a limited appreciation of the resources needed for implementation. The study proposed in this application will serve as a model for future development of standardized methods in cohort studies to return genomic results to study participants.









